
Synthesis of artificial proteins by combining
biological and chemical techniques
Laboratory on Nonnatural Protein Biosynthesis
Professor:HOHSAKA Takahiro
E-mail:
[Research areas]
Genetic engineering, Protein chemistry
[Keywords]
Genetic code expansion, Nonnatural amino acid, Artificial protein, Cell-free translation, Fluorescence analysis
Skills and background we are looking for in prospective students
Students should be interested in proteins and genes. Basic knowledge and experimental skills on biological chemistry and organic chemistry are required, but students will learn them after entering JAIST.
What you can expect to learn in this laboratory
Students can learn professional knowledge and experimental techniques on genetic engineering, protein synthesis, organic synthesis, fluorescence analysis, etc. Through research activities, students can learn a series of research processes ranging from planning of experiments, investigation of related researches, acquisition and analysis of experimental data, summary of research results, and presentation of achievements. These abilities are indispensable as both researchers and engineers.
【Job category of graduates】 Chemical or biological companies, research institutes
Research outline
Our aim is to create new artificial proteins by combining biochemical techniques such as genetic engineering and protein synthesis with chemical methods such as organic synthesis. Specifically, we are pursuing the following research themes. We are also conducting research to make practical use of our achievements with companies.
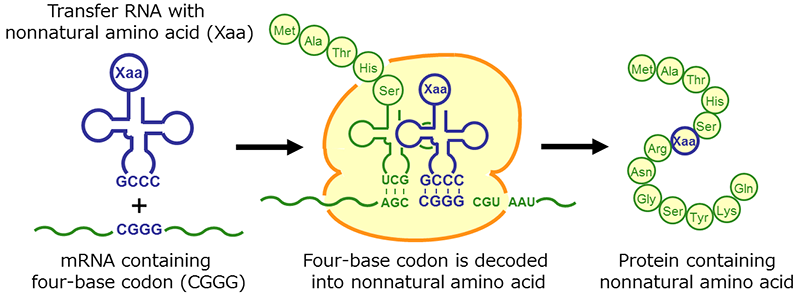
Fig. 1 Incorporation of nonnatural amino acids into proteins by using four-base codon.
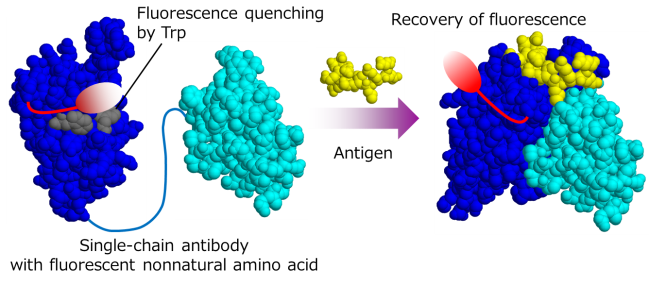
Fig. 2 Antibody-based fluorescence sensor.
1. Introduction of nonnatural amino acids into proteins by expansion of the genetic code
Proteins are made from amino acids according to the genetic code of DNA and show various biological functions by forming three-dimensional structures. However, only 20 types of amino acids are used in nature for protein biosynthesis. Beyond this limitation, we have succeeded in developing a new technology that can introduce artificially synthesized nonnatural amino acids into specific positions of proteins in response to expanded genetic code such as four-base codons (Fig.1).
2. Design and synthesis of artificial proteins
By using the above technology, we are working on design and function of artificial proteins with new functions. For example, protein sensors that can detect target molecules by fluorescence have been generated by introducing fluorescent nonnatural amino acids into ligand-binding proteins and antibodies (Fig.2). We are also tried synthesizing new protein or peptide drugs by utilizing the nonnatural amino acid incorporation technique. Parts of these studies are being in progress through collaboration with companies.
3. Development of new biotechnologies using potential of organisms
It can be said that the nonnatural amino acid incorporation technique is derived from the potential function of organisms. We are trying to develop another biotechnology that can synthesize useful artificial proteins by utilizing new potential of organisms.
Key publications
- Double-fluorescent-labeled single-chain antibodies showing antigen-dependent fluorescence ratio change, K. Yoshikoshi, T. Watanabe, T. Hohsaka, Bull. Chem. Soc. Jpn., 2016, 89, 573-580.
- Antibody-based fluorescent and fluorescent ratiometric indicators for detection of phosphotyrosine, K. P. Huynh Nhat, T. Watanabe, K. Yoshikoshi, T. Hohsaka, J. Biosci. Bioeng., 2016, 122, 146-154.
- "Quenchbodies": Quench-based antibody probes that show antigen-dependent fluorescence. R. Abe, H. Ohashi, I. Iijima, M. Ihara, H. Takagi, T. Hohsaka, H. Ueda, J. Am. Chem. Soc., 2011, 133, 17386-17394.
Equipment
DNA analysis (DNA sequencer, real-time PCR, etc.), Fluorescence analysis (Fluorescence spectrometer, Fluorescence scanner, Fluorescence microscope, etc.), Mass spectrometer, Analytical and preparative HPLCs, etc.
Teaching policy
Our goal is not only to obtain new knowledge from experiments but also to learn research process for artificial proteins. Specifically, we will train the ability to perform research such as planning the experiment, interpreting the result, solving the problem, feedback to the next experimental plan, etc. through repeated trial and error experiments. We regularly have research meetings for checking research status and providing guidance. We also have the opportunity to present the research results at academic conferences.
[Website] URL:https://www.jaist.ac.jp/ms/labs/hohsaka/