
Exploring structure-function relationship of proteins using NMR spectroscopy
Laboratory on Protein NMR
Professor:OHKI Shinya
E-mail:
[Research areas]
Protein NMR
[Keywords]
Protein, Structure-function relationship, NMR
Skills and background we are looking for in prospective students
To start the research work at my group, students should have at least one of the basic knowledge of biochemistry, biotechnology, bioinformatiocs and NMR at undergrad level. The students who are good at computer work are also welcome.
What you can expect to learn in this laboratory
The goal of our research work is understanding biological functions of proteins at molecular level based on their three-dimensional structures and dynamics. For this purpose, we are employing modern NMR techniques. My group is the super-user of four NMR machines (400MHz x 2, 500MHz, and 800MHz) in JAIST, thus the students in my group are expected to be NMR experts. Because we are working on biomolecules, a wide variety of skills for preparing and analyzing them will also be acquired. Scientific writing, presentation and discussion skills are of course trained at group meeting and conferences.
【Job category of graduates】 R & D of food, chemical, and pharmaceutical companies
Research outline
(1) Development of stable-isotope labeling methods
Protein NMR samples are necessary to be labeled with NMR active nuclei, such as 13C and 15N. Techniques for preparing such NMR samples are called as “stable-isotope labeling”. The labeling is normally achieved by using genetically modified E. coli., but not all protein samples can be expressed by E. coli. Thus, many other methods have been proposed. As one of the alternatives, we had developed a method to use plant BY2. Because BY2 is a higher cell, it has a potential to express complex proteins which are difficult for E. coli. to synthesize. Moreover, using BY2, we had optimized the protocol for labeling only Ile, Leu, and Val residues in the sample protein. We are continuously trying to develop some new labeling methods using the BY2 system.
(2) Proteins and peptides containing disulfide bonds
It is difficult for E. coli system to express proteins and peptides containing disulfide bonding (SS-bond). However, the BY2 system can do it. Thus, we have been trying to explore structure-function relationship of proteins and peptides containing SS-bonds. Here two topics what we had done are described briefly for your better understanding.
Stomagen, one of the peptide hormones, increases stomatal density of plants. Because stomagen contains three SS-bonds, sufficient amount sample was not prepared using E. coli system. We, however, could prepare the stable-isotope labeled stomagen using the BY2 system, and studied its structure and dynamics. The NMR results opened the discussion for understanding structure-function relationship of the family members at atomic resolution. Until today, we have successfully designed and actually prepared the mutants showing desired bioactivity to increase or decrease the stomatal density. It is believed that such basic science will contribute to solve recent issues on food yield and earth environment in the future.
ESF, containing four SS-bonds, acts at the early phase in seed development. When ESF is inactive, the size and shape of seeds become heterogeneous. We have solved the three-dimensional structure of ESF prepared by the BY2 system. And the structure has indicated the key residues for its activity. It is expected that controlling the ESF activity might provide many applications to harvest big seeds for agriculture and fruits without seeds for easy food processing, etc.
We are also handling various other proteins and peptides containing SS-bonds, such as defensins and toxins. Knowledge of their structure-function relationship will help for developing new drugs.
(3) Proteins in the signaling pathways
A wide variety of biological signals in cells is harmonized by proteins, lipids, nucleotides, etc. Post-transcription modification including phosphorylation and methylation is known as one of the cues for activating/inactivating the biological functions. The chemical modification is a trigger for structural change, resulting the new conformer interacting with the target molecule. We are focusing on monitoring the conformational transition of the proteins at work.
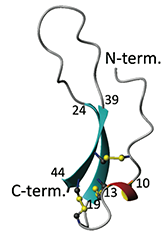
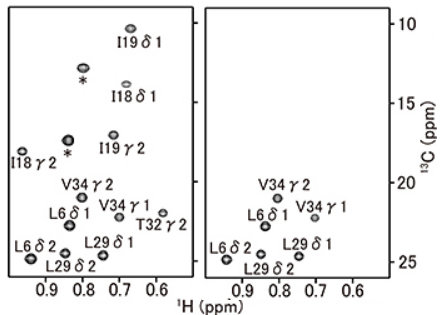
Figure. (left) Three-dimensional structure of stomagen.
(right) Methyl region of 1H-13C CT-HSQC of BPTI. Leu and Leu/Val selective labeling are achieved.
Key publications
- T. Imamura, H. Takagi, A. Miyazato, S. Ohki, H. Mizoguchi & M. Mori “Isolation and characterization of the betalein biosynthesis gene involved in hypocotyl pigmentation of the allotetraplois Chenopodium quinoa” Biophys. Biochem. Res. Commun. (2018) 496, 280-286.
- L.M. Costa, E. Marshall, M. Tesfaye, K.A.T. Silverstein, M. Mori, Y. Umetsu, S.L. Otterbach, R. Papareddy, H.G. Dickinson, K. Boutiller, K.A. VandenBosh, S. Ohki & J.F. Gutierrez-Marcos “Central cell-derived peptides regulate early embryo patterning in flowering plants” Science (2014) 344, 168-172.
- S. Ohki, M. Takeuchi & M. Mori “The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones” Nat. Commun. (2011) 2, 512.
Equipment
(in my group) deep-freezers, PCR, UV meter, centrifuges, bio-shakers, incubators, HPLCs, Linux machines, SPR, etc.
(open facility) 800MHz-NMR equipped with a TCI cryogenic probe, MALDI-TOF/TOF MS, FT-MS, etc.
Teaching policy
For the better scientific research work, discussion based on the experimental results is an undoubted important factor. Thus, students are recommended to discuss each other. And students can have opportunity to talk about his/her research progress at group meeting. Through the research, students get the knowledge how to plan small projects and to organize them toward the big goal.
[Website] URL:https://www.jaist.ac.jp/nmcenter/labs/s-ohki-www/contents/Ohki_Lab.html